A while ago, kaggle hosted the ultrasound nerve segmentation challenge, which requires partipants to predict the nerve area (brachial plexus) in a given Ultrasound image. The task is to predict the segmentation mask for the the brachial plexus. Even my own neural network (brain) finds it difficult to spot patterns in these images. Can you spot the pattern?
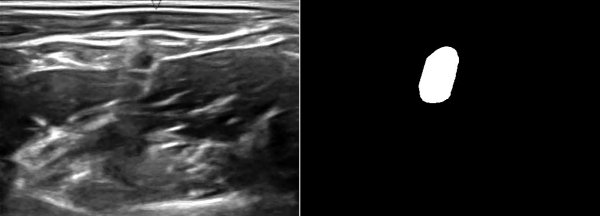
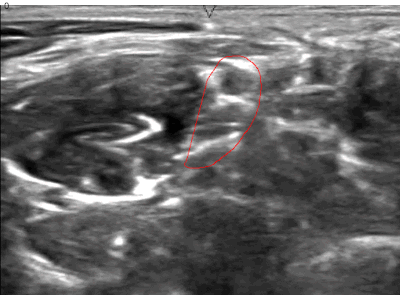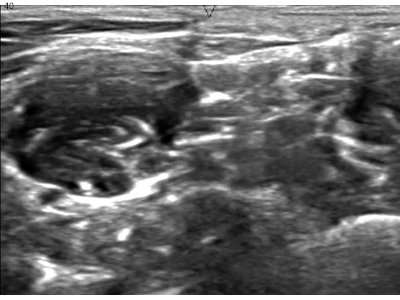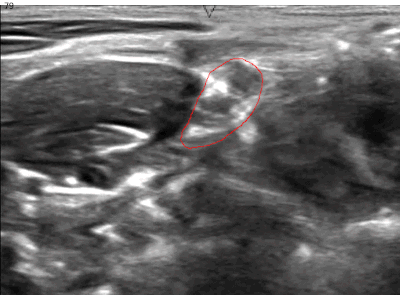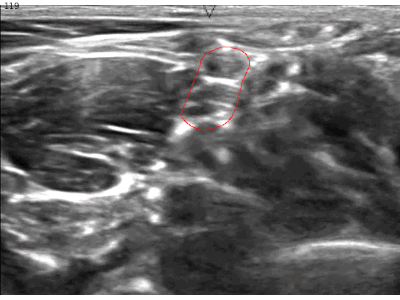
What makes this challenge particularly interesting is that ~60% of images do not contain the brachial plexus, i.e., segmentations mask is empty. Since the competition uses the mean of dice coefficient across all samples as the evaluation metric, even a single pixel in segmentation mask for non-nerve images mask kills the score. Ironically, you can achieve a score of ~0.6 just by predicting zero masks; it drops to ~[0.35, 0.45] when you try to learn it using a standard U-Net. This, combined with nosiy ultrasound images and a lack of obvious nerve patterns, intrigued me enough to participate in the challenge.
Unlike most kaggle posts, this article is more about the thought-process involved in coming up with a solution. I find it distasteful when folks just hand out a giant blob of code on github or just post their final complex solution that somehow gives good results.
Step1: Quick and dirty baseline
My methodology is always to first start with a quick and simple baseline model and setup and end-end working code. In case of this challenge, this means:
-
Data related I/O - Loading, train/test splits, optional preprocessing blocks
-
Baseline model: I stated with a U-Net as it seemed like the state of the art model for these kinds of data. I also retrofitted it with batch normalization, ELU activation, and dropout.
-
Clean code: Try to organize code into classes. Having function blocks everywhere makes the code messy and error prone.
-
Submission code: Code to generate submission file.
As mentioned in the overview, this tends to give mean dice score of ~[0.35, 0.45], which is pretty abysmal.
Step2: Data exploration
Now that an end-end working code is setup. We can work on optimizing individual blocks. The most obvious strategy is to get a better understanding of dataset at hand. The idea is to leverage insights from the dataset to improve/build the conv-net architecture and perhaps use dataset specific idiosynchronicities to provide better training signal.
Initial manual exploration revealed contradictory examples. i.e., two very similar images have different labels, one with a valid segmentation mask while the other doesn’t.
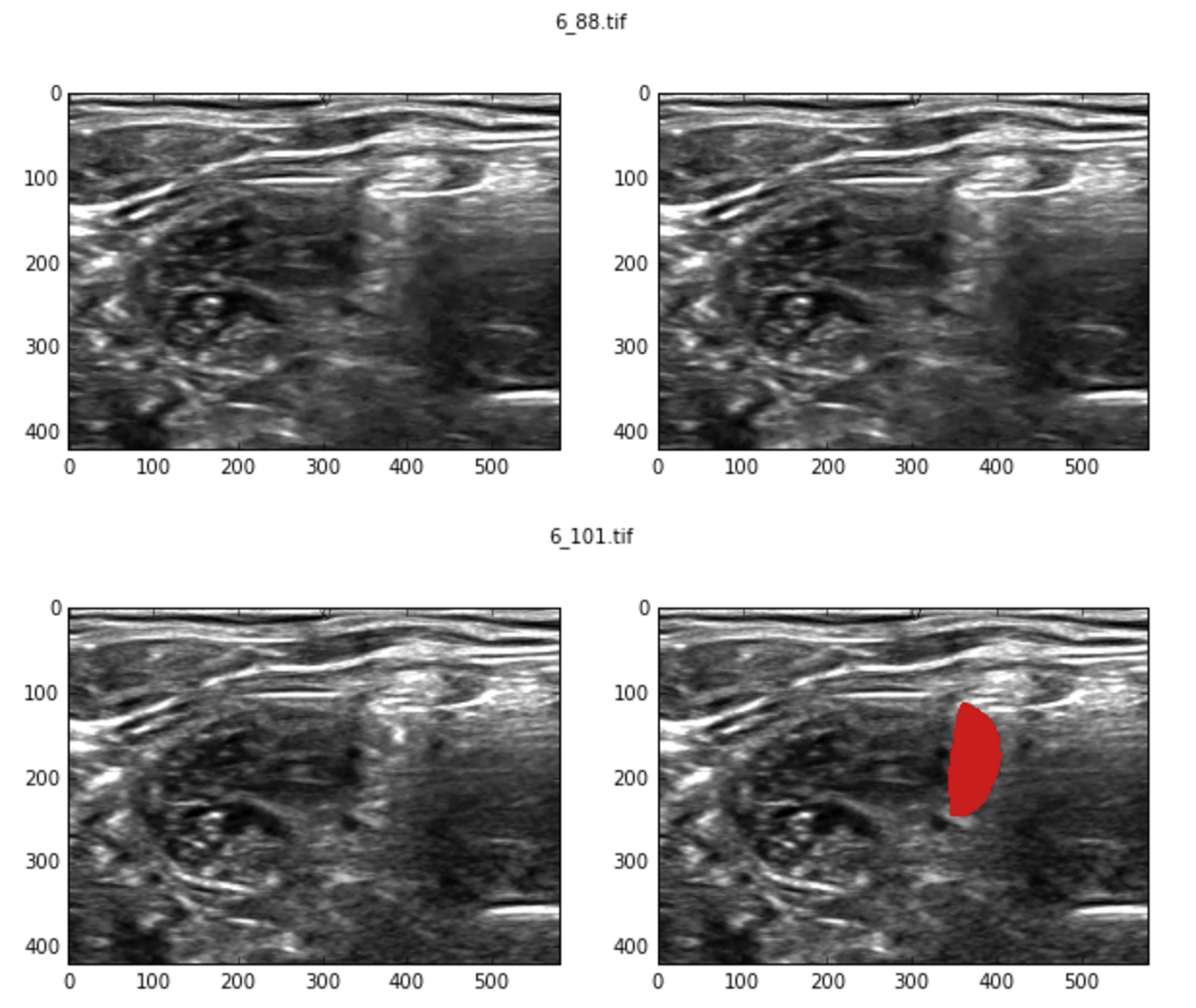
Lets try to find all such near duplicates. High level idea is as follows:
-
Divide image into blocks, say (20, 20).
-
Compute histogram over various blocks. The concatenated set of histograms represents the image hash.
-
Compute pairwise cosine similarity between image hashes and find ones within some reasonable threshold.
import skimage.util
import scipy.spatial.distance as spdist
def compute_img_hist(img):
# Divide the image in blocks and compute per-block histogram
blocks = skimage.util.view_as_blocks(img, block_shape=(20, 20))
img_hists = [np.histogram(block, bins=np.linspace(0, 1, 10))[0] for block in blocks]
return np.concatenate(img_hists)
# Compute hashes over all images
hashes = np.array(map(compute_img_hist, imgs))
# pairwise distance matrix
dist = spdist.squareform(spdist.pdist(hashes, metric='cosine'))
# find duplicates. +np.eye because we want to exclude image matching with itself.
# 0.008 is determined after some trial and errors.
close_pairs = dists + np.eye(dists.shape[0]) < 0.008The number of samples with inconsistent labeling is a whooping ~40% of the training set!!! Such a large number of inconsistent samples is bound to put an upper limit on the accuracy of the model. I think part of the problem lies with how the dataset was setup. Typically, doctors use ultrasound video frames to localize the nerve. Finding the nerve with a single frame is extremely challenging, even for human experts. I think it would be more interesting if they included videos instead.
The training set is small as is, just 5635 samples. After discards, we are only left with 3398 samples. This certainly doesn’t look good for training a deep model as they require lots of training data. On top of that, we cannot use transfer learning either as there are no pretrained-nets on this sort of data.
Despite all this, the cleaned raining data manages to improve baseline model score to ~0.55.
Step3: Real work begins…
| This article is still a work in progress. Check back for updates. |
Since the beginning, it was clear that i had to do something about the weird score evaluation. Even a single pixel predicted on the segmentation output for non-nerve images was going to hurt the score. A pretty straight forward strategy was to add a Dense layer towards the end of encoder an predict whether the image contains the mask or not. Lets call this value has_mask
During test/submission time, I could simply zero out all the segmentation mask values if has_mask = False.